A team of Bangladeshi scientists has achieved a significant milestone by generating the first-ever single-cell genomic data from Bangladesh for the Human Cell Atlas. This groundbreaking endeavor, backed by the Chan Zuckerberg Initiative, is part of the ‘Global Pediatric Cell Atlas of Nasal and Oral Mucosa’ project, conducted in collaboration with esteemed institutions such as Harvard University, MIT, and Boston Children’s Hospital. The primary objective of this initiative is to develop a pediatric atlas specifically focusing on the nasal and oral mucosa.
You Can Also Read: Brazil Releases Anti-Dengue Mosquitoes to Stem Outbreak: Will It Work?
To accomplish this goal, researchers at the Child Health Research Foundation (CHRF) are actively collecting samples from children residing in rural areas. Leveraging cutting-edge single-cell genomics technology, they aim to delve into the intricacies of individual human cells.
This initiative holds immense significance as it not only marks a significant diversification of genomic data sources beyond Western countries but also promises to contribute substantially to the development of more inclusive vaccine and therapy strategies.
‘Oral and nasal mucosa serve as the primary interfaces for environmental and microbial interactions but lack detailed data across diverse ancestries. This project will not only enhance our understanding of pediatric nasal and oral mucosa at a cellular level but also significantly contribute to Human Cell Atlas data from South Asia,’
– Dr. Senjuti on her verified Facebook account.

Dr. SenjutiSaha, Deputy Executive Director at CHRF, highlighted the significance of this achievement in enhancing our understanding of pediatric Nasal and Oral Mucosa and enriching the Human Cell Atlas with data from South Asia. This achievement underscores the collaborative efforts of Bangladeshi scientists in contributing to global scientific endeavors and advancing our understanding of human biology at a cellular level.
What is Single-Cell Genomics?
Single-cell genomics is a cutting-edge field that focuses on studying individual cells at a molecular level, providing detailed insights into their genetic and functional characteristics. This approach involves analyzing the nucleic acid sequence information of single cells, enabling a higher resolution of cellular differences and a deeper understanding of individual cell functions within complex biological systems. By examining the DNA or RNA of individual cells, researchers can uncover crucial information about mutations, gene expression patterns, and cellular behaviors that may not be apparent in bulk analyses.
The advent of single-cell genomics has revolutionized biological research by allowing scientists to explore cellular heterogeneity, track cell lineage relationships, and identify rare cell populations that play significant roles in various biological processes. This technology has been instrumental in diverse fields such as cancer research, developmental biology, and immunology. For instance, single-cell sequencing has been pivotal in characterizing different cell types during development, understanding myoblast differentiation, and unraveling immune cell populations in healthy and diseased states.
Moreover, single-cell genomics offers a powerful tool to investigate drug resistance mechanisms, disease progression, and cellular dynamics with unprecedented detail. By delving into the genetic and molecular profiles of individual cells, researchers can gain valuable insights into complex biological phenomena that were previously challenging to study comprehensively. Overall, single-cell genomics represents a transformative approach that holds immense potential for advancing our understanding of cellular biology and its implications for human health and disease.
Bioinformatics In Single Cell Genomics
In developmental biology, where the availability of a limited number of cells for analysis is common, the significance of single-cell analysis is heightened. Minute spatial and temporal variations can trigger significant changes in cell behavior during the intricate differentiation process. The experimental phase of single-cell analysis is typically segmented into three key stages: (i) employing biotechnological and microfluidics techniques, (ii) utilizing various platforms such as imaging, sequencing, microarray, and spectrometry for data acquisition, and (iii) conducting data analysis.
Imaging has played a pivotal role in cell biology progress since the advent of the microscope, enabling the visualization of proteins and cellular components. Despite its reliability challenges, antibody staining remains the conventional method for protein observation in fixed cells.
Data analysis, particularly normalization, is crucial in single-cell bioinformatics due to amplification biases arising from limited RNA/DNA material. Addressing this issue necessitates a combination of experimental and computational strategies.
In single-cell sequencing projects, genomic and/or transcriptomic data from numerous individual cells are obtained. Comparative analysis aims to identify abundant structural variants or transcripts across different cells or cell populations, facilitating a deeper understanding of cellular heterogeneity.
Low-dimensional analysis is another essential aspect of single-cell genomics, focusing on identifying prevalent structural variants or transcripts within diverse cell populations, thereby enhancing our comprehension of cellular dynamics and diversity.
Applications of Single-Cell Genomics
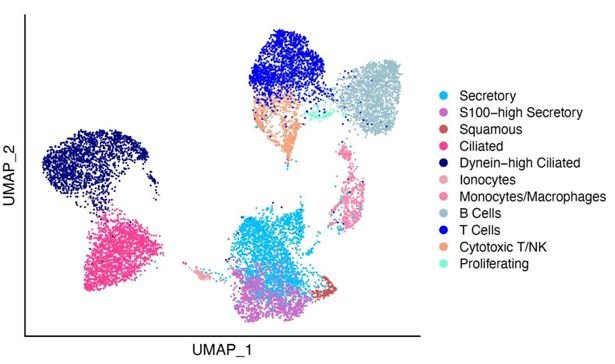
The application of single-cell RNA sequencing scRNA-seq (Figure 1) is revolutionizing our understanding of biology, expanding research horizons beyond mere cell state characterization. This advanced technology holds great promise for diverse medical applications. Tumor heterogeneity, a common feature within and among tumors, can greatly benefit from scRNA-seq, revealing previously unknown tumor characteristics that may have been overlooked in bulk transcriptomic analyses. By harnessing scRNA-seq, researchers can shed light on hidden aspects of tumor biology, potentially advancing the development of cancer evolution models.

Moreover, by modeling transcriptional kinetics, this method has the potential to reconstruct clonal and phylogenetic relationships among cells, offering a deeper insight into cellular dynamics and interactions.
Another Bangladeshi scientist
Another Bangladeshi scientist has been named to the 2023 list of the “best and brightest” 100 Asian scientists for their research contributions. In Asia, researchers are continuing to pursue ambitious goals while actively serving marginalized communities.
Among the notable achievers is Dr. GawsiaWahidunnessa Chowdhury, recognized for her significant contributions to sustainability.
Dr. Chowdhury, a distinguished zoology professor at the University of Dhaka, holds a PhD in zoology (wetland ecology) from the University of Cambridge. She also serves on the board of the esteemed conservation organization, WildTeam, dedicated to preserving Bangladesh’s rapidly depleting natural resources.
In 2022, Dr. Chowdhury was honored with the OWSD-Elsevier Foundation Award for her outstanding efforts in conserving aquatic environments and protecting vulnerable species in Bangladesh. Her concerns extend to the threat of plastic pollution in the country’s water bodies.
Conclusion
The pioneering efforts of Bangladeshi scientists in spearheading this single-cell genomic revolution underscore the transformative power of collaborative research and innovation. As the scientific community celebrates this historic milestone, the journey towards unlocking the mysteries of human biology at a cellular level continues, promising to usher in a new era of discovery and advancement in biomedical science.


